CoGNaC: A Chaste Plugin for the Multiscale Simulation of Gene Regulatory Networks Driving the Spatial Dynamics of Tissues and Cancer
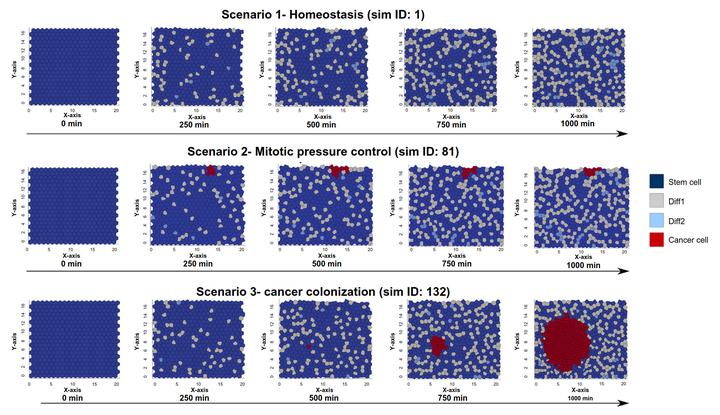 Image credit: SAGE
Image credit: SAGE
Abstract
We introduce a Chaste plugin for the generation and the simulation of Gene Regulatory Networks (GRNs) in multiscale models of multicellular systems. Chaste is a widely used and versatile computational framework for the multiscale modeling and simulation of multicellular biological systems. The plugin, named CoGNaC (Chaste and Gene Networks for Cancer), allows the linking of the regulatory dynamics to key properties of the cell cycle and of the differentiation process in populations of cells, which can subsequently be modeled using different spatial modeling scenarios. The approach of CoGNaC focuses on the emergent dynamical behavior of gene networks, in terms of gene activation patterns characterizing the different cellular phenotypes of real cells and, especially, on the overall robustness to perturbations and biological noise. The integration of this approach within Chaste’s modular simulation framework provides a powerful tool to model multicellular systems, possibly allowing for the formulation of novel hypotheses on gene regulation, cell differentiation, and, in particular, cancer emergence and development. In order to demonstrate the usefulness of CoGNaC over a range of modeling paradigms, two example applications are presented. The first of these concerns the characterization of the gene activation patterns of human T-helper cells. The second example is a multiscale simulation of a simplified intestinal crypt, in which, given certain conditions, tumor cells can emerge and colonize the tissue.